Journal of Cardiovascular Medicine and Cardiology
New cases of rare dyslipidemias in clinical practice
C Rodríguez-Jiménez1,2#, J Sanguino1,2#, E Sevilla-Alonso1,2, F Arrieta3, I García-Polo4, JM Mostaza5 and S Rodríguez-Nóvoa1,2*
2Group of Dislipemias of Genetic Origin and Metabolic Diseases, IdiPAZ, La Paz University Hospital, Madrid, Spain
3Department of Endocrinology & Nutrition, Hospital Universitario Ramón & Cajal, Group Metabolic Disease, IRYCIS, Spain
4Department of Internal Medicine, Princess Hospital, Madrid, Spain
5Internal Medicine Service, Hospital Carlos III, HULP, Madrid, Spain
#These authors contributed equally to this work
Cite this as
Rodríguez-Jiménez C, Sanguino J, Sevilla-Alonso E, Arrieta F, García-Polo I, et al. (2022) New cases of rare dyslipidemias in clinical practice. J Cardiovasc Med Cardiol 9(4): 030-036. DOI: 10.17352/2455-2976.000185Copyright License
© 2022 Rodríguez-Jiménez C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Lipid metabolism can experience different disorders resulting in changes in the function and concentrations of plasma lipoproteins. These changes affect alone or interact with other cardiovascular risk factors involved in the development of atherosclerosis. Therefore, dyslipidemias cover a wide spectrum of disorders lipids. Some of them have a genetic origin and very low prevalence. The main objective of this article is to report new cases of rare dislipemias of genetic origin in our population.
Genetic analysis was performed by Next Generation Sequencing (NGS) using a customized panel of 436 genes in DNA samples of four patients. The results confirmed the genetic origin of the following dyslipidemias: fish-eye disease, primary hypoalphalipoproteinemia-2, familial hypercholesterolemia by a variant in STAP1 and Sitosterolemia.
This approach allows us to confirm the genetic diagnosis of four patients with alterations in lipid metabolism, this will help to improve patient management, achieving early diagnosis in the study of family members
Introduction
Dyslipidemias are alterations in lipid metabolism that occur with variations in lipid concentrations. Different studies have shown that modifications in levels of plasma lipoproteins are associated with the development of cardiovascular disease [1-3].
In some cases, alterations in lipid levels can be deter mined by genetic factors. So far, different monogenic lipid disorders with low prevalence had been identified [4], such are Fish-eye disease (FED; MIM# 136120), Primary hypoalphalipoproteinemia-2 (MIM#618463), Sitosterolemia (STSL1; MIM #210250) and Familial Hypercholesterolemia (FH; MIM#143890) caused by variants in STAP1 among others.
Fish-eye disease is a rare disease; approximately 30 cases have been reported in the medical literature, with an inheritance pattern autosomal recessive caused by partial Lecithin Cholesterol Acyltransferase deficiency (LCAT ) [5]. The LCAT enzyme plays a central role in removing cholesterol from the blood and tissues. This soluble enzyme converts cholesterol and phosphatidylcholines lecithins to cholesteryl esters and lysophosphatidylcholines on the surface of High-Density Lipoproteins (HDL). Two activities are related to LCAT , the main is alpha LCAT activity in HDL, furthermore has beta LCAT activity in Very Low-Density Lipoproteins (VLDL) and Low-Density Lipoproteins (LDL). Pathogenic variants at the LCAT gene can cause fish-eye disease impairing alpha LCAT activity, therefore, reducing the enzyme’s ability to attach cholesterol to HDL. This disorder is characterized by increasing cloudiness of the corneas in the patients due to small grayish dots of cholesterol in them that can lead to severely impaired vision and very low levels of HDL-cholesterol (HDL-c). Other typical findings are raised levels of total cholesterol, and triglycerides and decreased levels of Apolipoprotein A-I (apoA-I) and Apolipoprotein B (apo B) in serum.
Primary hypoalphalipoproteinemia-2 is generally an autosomal recessive disorder associated with extremely low levels of apoA-I and HDL-c in serum, xanthomas, corneal opacities and in most patients, premature atherosclerotic cardiovascular disease [6]. Currently, 30 families have been described. This disease is caused by pathogenic variants in the APOA1 gene (MIM 107680) that code for the apoA-I, the major structural component of HDL. It is also needed to activate LCAT and to mediate the interaction of HDL with cell surface receptors, such as scavenger receptor B1, or plasma membrane transporters such as ABCA1 [6].
Autosomal-dominant hypercholesterolemia is a common genetic disorder (1:250-1:500) [7], associated with extremely high levels of LDL-cholesterol (LDL-c). Molecular diagnosis can be confirmed by the presence of pathogenic variants in LDLRAP1 (low-density lipoprotein receptor, OMIM#606945), APOB (Apolipoprotein B, OMIM#107730), and PCSK9 (proprotein convertase subtilisin/Kexin type 9, OMIM#607786). Recent studies suggested the STAP1 (signal transducing adaptor family member 1, OMIM#604298) as the fourth FH gene [8].
Sitosterolemia is a rare autosomal recessive hereditary lipid storage disorder characterized by increased plant sterol levels, xanthomas and accelerated atherosclerosis [9]. It is caused by homozygous or compounds heterozygous mutations in one of the two ABCG5 (STSL2; MIM# 618666) and ABCG8 (STSL1; MIM #210250) genes encoding the sterol efflux transporter ABCG5 and ABCG8 . Recent studies have shown that the prevalence of subjects with deleterious variants in ABCG5 and/or ABCG8 genes could be more than 1 in ~200,000 individuals among the general population [10].
The main objective of this article is to report new cases of rare dyslipidemias of genetic origin in our population.
Materials and methods
Patients
The patients were referred to our Laboratory of Genetics of Metabolic diseases. These patients came from the Departments of Endocrinology & Nutrition and Internal Medicine. All patients were informed of the genetic tests performed and signed the informed consent. Clinical characteristics of patients are described individually in each of the cases presented throughout this work.
Genetic analysis
The genomic DNA (gDNA) from probands was extracted from EDTA-treated whole blood samples using Chemagen (Chemagic DNA extraction special, Perkin Elmer Inc, Baesweiler, Germany). DNA quantification was performed using a fluorimeter Invitrogen Qubit 2.0. Genetic analysis was performed by Next Generation Sequencing (NGS) using a customized panel of 500 genes. Library preparation and exome enrichment steps were performed according to the manufacturer´s workflow (Nimblegen, Roche) and it was sequenced using HiSeq4000 system Sequencing, Illumina. NGS data was suitable for analysis after passing the quality parameters established in our laboratory: a number of reads more than 30× in the 99% of the target bases. Sanger sequencing was used to confirm the presence of the new variants.
In silico analysis
Bioinformatic analysis was performed using algorithms developed by our bioinformatics unit. Briefly, sequences were mapped to the CRCh37/hg19 human reference sequence and databases used for analysis were Human Gene Mutation Database (HGMD®) (http://www.hgmd.cf.ac.uk/ac/index.php) from BIOBASE Corporation; Online Mendelian Inheritance in Man (www.omim.org); Gene Tests (www.genetests.org). Variant annotation was carried out with Ensembl´s variant Effect Predictor Tool and was based on the transcripts ENST00000264005.5-LCAT , ENST00000236850.4-APOA1 , ENST 00000558518-LDLRAP1 , ENST0000023324-APOB, ENST00000 302118-PCSK9 and ENST00000374338-LDLRAP1 AP1, ENST00000 265404.2-STAP1 , ENST00000260645.1-ABCG5 , ENST00000 272286.2-ABCG8 In silico predictors of pathogenicity used were CADD (Combined Annotation Dependent Depletion; Damaging|s>=14), Polyphen (Polymorphism Phenotyping; Benign|s<0.03), MutAssesor (Benign|s<1.12), Fasthmm (Damaging|s<=-1) and Vest (Benign|s<0.17). Scores of conservation used: Gerp2 (Conserved|s>2.45), PhasCons, Phylop in relation to thirteen species. MaxEntScan, NNSplice, GeneSplicer and Human Splicing Finder were used as splicing predictors. The factors taken into consideration were Damaging or Possibly Damaging to three or more pathogenicity predictors and conservation for Gerp2. The interpretation of sequence variants was carried out according to the recommendations of The American College of Medical Genetics and Genomics (ACMG) [11]. The files were uploaded in BAM format for analysis using Alamut Visual V.2.8.0 (Interactive Biosoftware; France).
Results
Results
A 27-years-old male with mixed dyslipidemia showed “white halo” in eye. He had familial antecedent of corneal opacity (father and grandfather), without loss of seeing. The patient was diagnosed of corneal lipoid dystrophy in fish-eye with prominent gerontoxon in the ophthalmology unit.
Complete biochemical analysis was performed, highlighting a very low level of HDL-c and raised levels of triglycerides, liver enzymes and homocysteine levels in serum (Table 1).
Hematologic and coagulation tests were normal. Serology about HCV, CMV, HIV, EBV and toxoplasmosis were negatives. Splenomegaly with normal liver without anemia or kidney disease was observed.
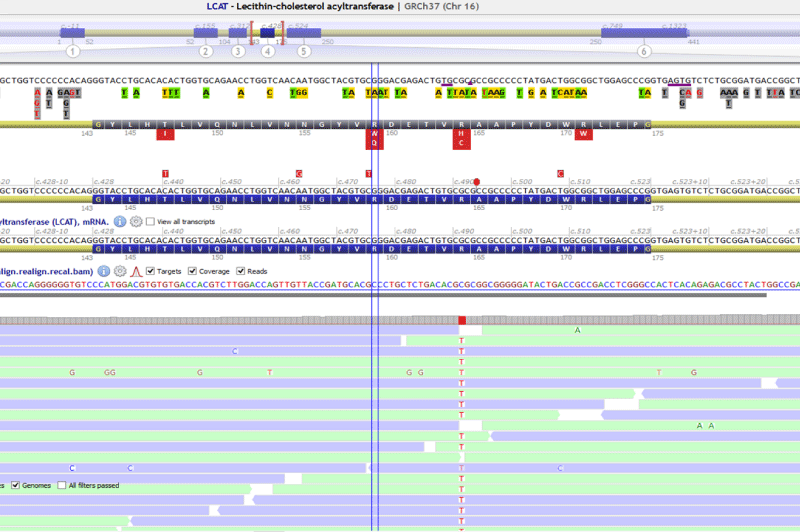
A suspicion of fish-eye disease was supposed due to clinical and biochemical findings. Differential diagnosis included Schnyder corneal dystrophy, familial LCAT deficiency and Tangier disease.
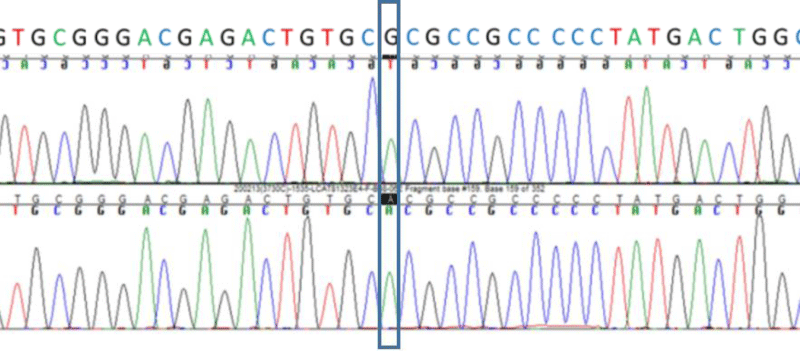
A missense variant in LCAT NM_000229.2:c.491G>A p.(Arg164His) was found an homozygous and confirmed by Sanger sequencing (Figure 1). In silico analysis showed a very low allelic frequency, high conservation and the bioinformatics prediction tools classified the variant as damaging (Table 2). No pathogenic variants were found in ABCA1. It was also found a common polymorphism in MTHFR, NM_005957.4:c.665C>T; (p.Ala222Val), that is related with hyperhomocysteinemia.
Case 2
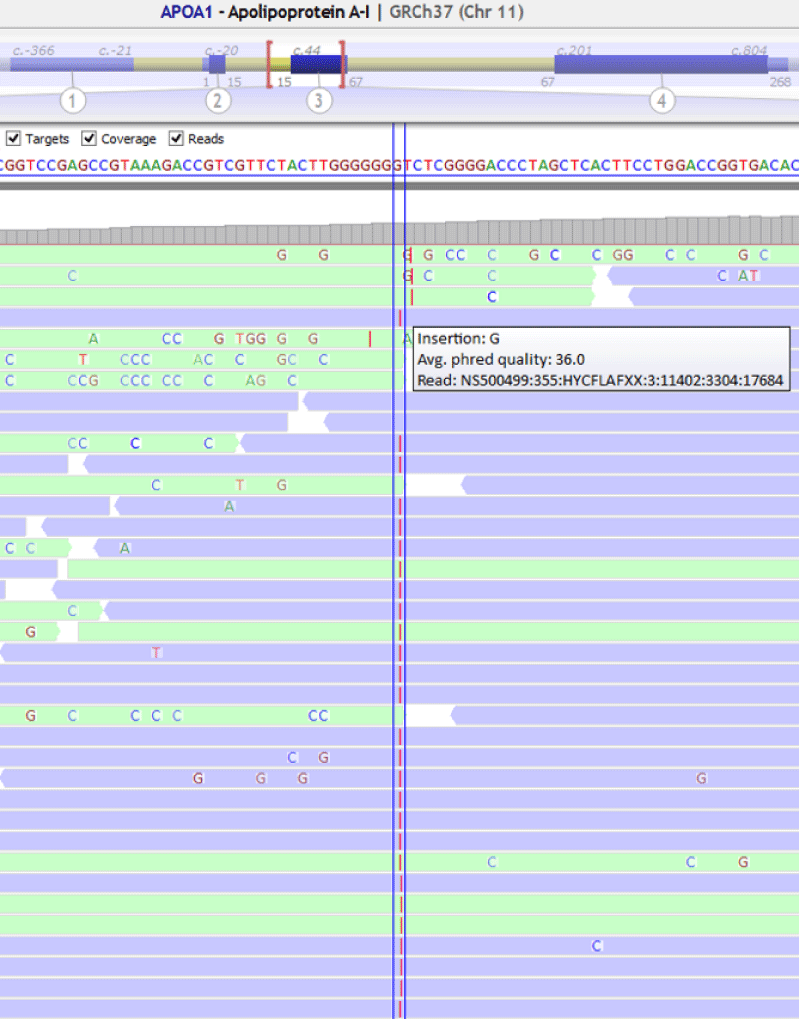
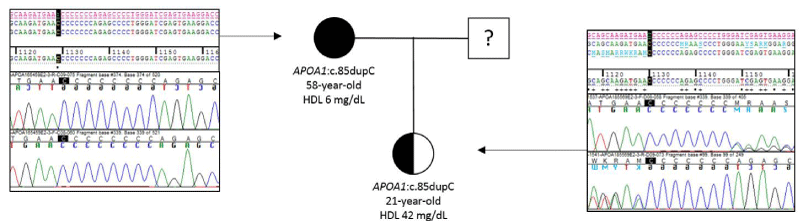
A 58-year-old woman with xanthomas, corneal clouding and paternal and maternal antecedents of isquemic cardiopathy was remitted to our service for genetic testing. The patient had extremely low levels of apoA-I (<16 mg/dL) and HDL-c (6 mg/dL) in serum (Table 3). We also tested the patient’s daughter, a 21-year-old woman with xanthomas, low levels of apoA-I (99 mg/dL) and normal levels of HDL-c (42 mg/dL) in serum. Sanger sequencing was used to confirm the familiar variant in APOA1 Figure 2.
The genetic analysis of the patient showed a frameshift variation in homozygosity in APOA1 , NM_000039.2:c.85dupC ;p.(Gln29Profs*29) (Figure 3). This variant was confirmed by Sanger sequencing. The analysis of the patient’s daughter showed the variant in APOA1 , NM_000039.2:c.85dupC ;p.(Gln29Profs*29) in heterozygous (Figure 4).
Case 3
Three patients remitted to our unit with suspicious of familiar hypercholesterolemia (FH) showed no pathogenic variants at the LDLRAP1 , APOB, PCSK9 and LDLRAP1 AP1. Characteristics of patients are shown in Table 4. In the extended analysis to genes related with lipid metabolism, we found variants in heterozygous at the STAP1 gene in each patient. Three variants have been previously reported.
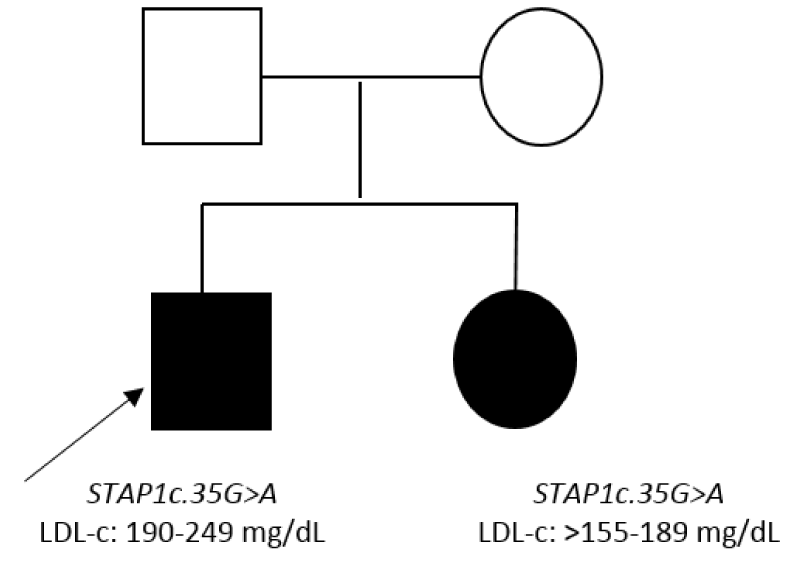
Patient 3.1 carried the variant STAP1 :NM_012108.3:c.35G>A:p.(Arg12His) and patient 3.2 carried the STAP1 :NM_ 012108.3:c.526C>T :p.(Pro176Ser), these variants showed frequencies less than 0.03%, were conserved and the in silicopredictors showed damage. Patient 3.3 carried the variant STAP1 :NM_012108.3:c.120+6T>C , this variant did not show impact according to splicing predictors (Table 2). The variant c.35G>A was found in the sister of patient 3.1presenting LDL-c>155 mg/dL (Figure 5). In the rest of the patients we were unable to carry out segregation studies because we did not get a sample.
Case 4
A 55-year-old woman with hypercholesterolemia total cholesterol 282 mg/dl, LDL-c 194 mg/dl and HDL-c 76 mg/dl, normal triglycerides 56 mg/dl , lp(a) 10.2 mg/dl and homocysteinemia 10.5 mg/dl Table 5. The patient have familial antecedent of vascular disease, mother with ischemic heart disease. The patients were being treated with statin and poor control, being referred for genetic study. The result of the genetic study of FH including the standard analysis of genes LDLRAP1 , APOB, PCSK9 and LDLRAP1 AP1 showed no pathogenic variants. In the extended analysis to other genes involved in lipid metabolism we found two variants in heterozygous in ABCG8 _NM_022437.2: c.788G>A;p.(Arg263Gln) and c.1083G>A;p.(Trp361*), both showed impact according to bioinformatic tools (Table 2). The variant p. (Arg263Gln) is a missense-type variant and the second variant, p. (Trp361 *) is a stop variant (null allele), both of them have been described in the literature associated with sitosterolemia [12-14].
Discussion
In this work, we have reported four cases of rare dyslipidemias. The genetic analysis help in final confirmation of the diagnosis. The variant found in LCAT c.491G>A;p.(Arg164His) had been previously reported in three siblings with typical triad of Familial Deficiency of LCAT : corneal opacity, hemolytic anemia and kidney dysfunction. Functional study showed complete deficiency of the LCAT enzyme [15]. However, our patient exhibited a mild phenotype despite of display of same variant. In other variants of LCAT , different clinical and biochemical pictures have been found among family members and suggesting the existence of others factors as environmental factors or other minor genes that may be involved in the clinical presentation of each patient.
The variant APOA1 : c.85dupC;p.(Gln29Profs*29) had been previously described as an autosomal dominant cause of apoA-I deficiency [16]. ApoA-I deficiency is generally associated with markedly increased risk of atherosclerotic cardiovascular disease. However, in this study we identified a frameshift mutation of the APOA1 gene as the molecular basis of apoA-I deficiency in a 58-year-old woman without isquemic cardiopathy. Patient’s daughter showed the variant in heterozygous without alteration neither the lipid profile or in clinical signs confirming cosegregation in this variant.
There is controversy about the role of STAP1 in FH [17]. Some studies have showed lack of cosegregation in some variants found in STAP1 in FH patients [18]. In this study we found three variants previously described in patients with hypercholesterolemia. The variant c.120+6T>C did not show impact; the variant c.526C>T :p.(Pro176Ser) was found in a patient but we could not analyze relatives and the variant c.35G>A:p.(Arg12His) was in a patient and his daughter. Further studies of cosegregation and functional studies should be performed in order to confirm the role of STAP1 in FH.
To date an estimated 30 variants have been described in each of the genes ABCG5 and ABCG8 associated with sitosterolemia (The Human Gene Mutation Database) [19]. According to The Exome Aggregation Consortium (ExAC) exome browser, 1 in ~220 individuals show loss of function (LOF) mutations in ABCG5 or ABCG8 genes [10]. Nevertheless, the prevalence of Sitosterolemia is under 1/1.000.000 indicating that could be underdiagnosed. Quantification of phytosteroles in our patients could be useful but we have not this possibility, however we were able to analyze the genes responsible for sitosterolemia and thus to contribute to the diagnosis of this disease.
There is no doubt that the genetic analysis using customized panels of genes related with metabolism of lipids is a useful tool to confirm or even to detect low frequency dislipidemias. In this work, we have reported four dyslipidemias of genetic origin due to variants of low frequency confirming the genetic diagnosis in these patients. The importance of reporting these cases has an impact on appropriate patient’s management as they allow an early diagnosis.
Financial support
The study was supported by grant FIS18/00917, FEDER, ISCIII-Instituto de Salud Carlos III, Spain.
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA. 2001 May 16;285(19):2486-97. doi: 10.1001/jama.285.19.2486. PMID: 11368702.
- Nelson RH. Hyperlipidemia as a risk factor for cardiovascular disease. Prim Care. 2013 Mar;40(1):195-211. doi: 10.1016/j.pop.2012.11.003. Epub 2012 Dec 4. PMID: 23402469; PMCID: PMC3572442.
- Adhyaru BB, Jacobson TA. New Cholesterol Guidelines for the Management of Atherosclerotic Cardiovascular Disease Risk: A Comparison of the 2013 American College of Cardiology/American Heart Association Cholesterol Guidelines with the 2014 National Lipid Association Recommendations for Patient-Centered Management of Dyslipidemia. Endocrinol Metab Clin North Am. 2016 Mar;45(1):17-37. doi: 10.1016/j.ecl.2015.09.002. PMID: 26892995.
- Authors/Task Force Members; ESC Committee for Practice Guidelines (CPG); ESC National Cardiac Societies. 2019 ESC/EAS guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Atherosclerosis. 2019 Nov;290:140-205. doi: 10.1016/j.atherosclerosis.2019.08.014. Epub 2019 Aug 31. Erratum in: Atherosclerosis. 2020 Jan;292:160-162. Erratum in: Atherosclerosis. 2020 Feb;294:80-82. PMID: 31591002.
- Santamarina Fojo S, Hoeg JM, Assmann G, Brewer HB. Lecithin Cholesterol Acyltransferase Deficiency and Fish Eye Disease. In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The Metabolic and Molecular Bases of Inherited Diseases. New York: McGraw Hill; 2001: 2817-2833.
- von Eckardstein A. Differential diagnosis of familial high density lipoprotein deficiency syndromes. Atherosclerosis. 2006 Jun;186(2):231-9. doi: 10.1016/j.atherosclerosis.2005.10.033. Epub 2005 Dec 15. PMID: 16343506.
- Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, Wiklund O, Hegele RA, Raal FJ, Defesche JC, Wiegman A, Santos RD, Watts GF, Parhofer KG, Hovingh GK, Kovanen PT, Boileau C, Averna M, Borén J, Bruckert E, Catapano AL, Kuivenhoven JA, Pajukanta P, Ray K, Stalenhoef AF, Stroes E, Taskinen MR, Tybjærg-Hansen A; European Atherosclerosis Society Consensus Panel. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013 Dec;34(45):3478-90a. doi: 10.1093/eurheartj/eht273. Epub 2013 Aug 15. Erratum in: Eur Heart J. 2020 Dec 14;41(47):4517. PMID: 23956253; PMCID: PMC3844152.
- Fouchier SW, Dallinga-Thie GM, Meijers JC, Zelcer N, Kastelein JJ, Defesche JC, Hovingh GK. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ Res. 2014 Aug 29;115(6):552-5. doi: 10.1161/CIRCRESAHA.115.304660. Epub 2014 Jul 17. PMID: 25035151.
- Escolà-Gil JC, Quesada H, Julve J, Martín-Campos JM, Cedó L, Blanco-Vaca F. Sitosterolemia: diagnosis, investigation, and management. Curr Atheroscler Rep. 2014 Jul;16(7):424. doi: 10.1007/s11883-014-0424-2. PMID: 24821603.
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug 18;536(7616):285-91. doi: 10.1038/nature19057. PMID: 27535533; PMCID: PMC5018207.
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24. doi: 10.1038/gim.2015.30. Epub 2015 Mar 5. PMID: 25741868; PMCID: PMC4544753.
- Hansel B, Carrié A, Brun-Druc N, Leclert G, Chantepie S, Coiffard AS, Kahn JF, Chapman MJ, Bruckert E. Premature atherosclerosis is not systematic in phytosterolemic patients: severe hypercholesterolemia as a confounding factor in five subjects. Atherosclerosis. 2014 May;234(1):162-8. doi: 10.1016/j.atherosclerosis.2014.02.030. Epub 2014 Mar 11. PMID: 24657386.
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000 Dec 1;290(5497):1771-5. doi: 10.1126/science.290.5497.1771. PMID: 11099417.
- Heimerl S, Langmann T, Moehle C, Mauerer R, Dean M, Beil FU, von Bergmann K, Schmitz G. Mutations in the human ATP-binding cassette transporters ABCG5 and ABCG8 in sitosterolemia. Hum Mutat. 2002 Aug;20(2):151. doi: 10.1002/humu.9047. PMID: 12124998.
- Steyrer E, Haubenwallner S, Hörl G, Giessauf W, Kostner GM, Zechner R. A single G to A nucleotide transition in exon IV of the lecithin: cholesterol acyltransferase (LCAT ) gene results in an Arg140 to His substitution and causes LCAT -deficiency. Hum Genet. 1995 Jul;96(1):105-9. doi: 10.1007/BF00214196. PMID: 7607641.
- Nakata K, Kobayashi K, Yanagi H, Shimakura Y, Tsuchiya S, Arinami T, Hamaguchi H. Autosomal dominant hypoalphalipoproteinemia due to a completely defective apolipoprotein A-I gene. Biochem Biophys Res Commun. 1993 Oct 29;196(2):950-5. doi: 10.1006/bbrc.1993.2341. PMID: 8240372.
- Danyel M, Ott CE, Grenkowitz T, Salewsky B, Hicks AA, Fuchsberger C, Steinhagen-Thiessen E, Bobbert T, Kassner U, Demuth I. Evaluation of the role of STAP1 in Familial Hypercholesterolemia. Sci Rep. 2019 Aug 19;9(1):11995. doi: 10.1038/s41598-019-48402-y. PMID: 31427613; PMCID: PMC6700100.
- Lamiquiz-Moneo I, Restrepo-Córdoba MA, Mateo-Gallego R, Bea AM, Del Pino Alberiche-Ruano M, García-Pavía P, Cenarro A, Martín C, Civeira F, Sánchez-Hernández RM. Predicted pathogenic mutations in STAP1 are not associated with clinically defined familial hypercholesterolemia. Atherosclerosis. 2020 Jan;292:143-151. doi: 10.1016/j.atherosclerosis.2019.11.025. Epub 2019 Nov 29. PMID: 31809983.
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003 Jun;21(6):577-81. doi: 10.1002/humu.10212. PMID: 12754702.